Antibodies (Ab), also known as immunoglobulins (Ig), are large, Y-shaped proteins that help the immune system recognize and destroy foreign substances like bacteria and viruses. The antigen, which is a unique molecule of the pathogen, is recognized by the antibody.
Antibody conjugation methods include reactions with amine side chains on lysine amino acids, disulfide rebridging, using N-hydroxysuccinimide to attach fluorophores, using EDC-DMAP for attaching nanoparticles, and many more.
What is a Conjugated Antibody vs. Unconjugated Antibody?
The difference between a conjugated antibody vs. unconjugated antibody is that a conjugated antibody has been connected to a substrate such as a toxin, enzyme, drug, or inorganic molecule while an unconjugated antibody has not been connected to a substrate.
Antibodies that have been conjugated are also called (also called a labeled, tagged, or loaded antibody) and are used in a variety of scientific applications, ranging from Western blotting to in vivo cellular investigation (source). These labeled antibodies can be used to separate and purify a desired protein from a complicated mixture, such as cells, tissues, or complete organisms.
Most proteins and antibodies have readily available amines and carboxyls. You can label proteins and antibodies with these conjugation kits to save time and improve consistency between experiments.
Overview of Antibody Conjugation Methods
Antibody conjugation is normally done chemically, however specialized enzymatic conjugation, such as sortase-based protein terminal conjugation or even protein engineering, is becoming more prevalent. The –NH2 (amino) group of a lysine or the free –SH (sulfhydryl) group of cysteine are the favored sites for chemical conjugation on an antibody.
Antibody-Drug Conjugation Methods
Antibody-drug ADCs, or antibody-drug conjugates, are highly targeted biopharmaceutical therapies that combine monoclonal antibodies specific to surface antigens found on specific tumor cells with highly effective anti-cancer medications linked via a chemical linker. The ability of antibody-drug conjugates to selectively deliver very potent cytotoxic anticancer chemotherapies directly to cancer cells while leaving healthy tissue unharmed is a unique attribute of these so-called armed antibodies (source). For a more specific example of antibody-drug conjugation, check out our article about bifunctional chelating agents and how we use them.
Antibody-drug conjugation methods typically involve attachment to lysine groups or disulfide-rebridging, which involves reducing the disulfide links and then introducing a linker that re-bridges the reduced thioethers and provides a location to attach a drug.
A monoclonal antibody, a stable linker, and a cytotoxic substance are the main components of antibody-drug conjugates.

An easy technique to create antibody-drug conjugates is to attach the drug to lysine groups on the antibody since the amine side-chains are highly reactive. However, this technique leads to polydisperse samples since there are so many lysine groups on the antibody protein. You can learn more about lysine conjugation protocols in our related article.
Disulfide rebridging with a site-selective approach has emerged as one major technique for producing homogenous ADCs. Here, interchain disulfides are first reduced and then treated with a linker which crosslinks the reactive thiols from the reduced disulfides. This reconstructs the covalent bridge between the protein chains and introduces a location for a drug to be attached.
Bissulfones, next-generation maleimides, and pyridazinediones are currently among the reagents available for disulfide rebridging linkers, which have made significant progress in recent years. Despite the fact that these platforms represent important advancements, new techniques are needed to accelerate the creation of homogenous ADCs including native antibodies (source). We’ve covered protocols utilizing bioconjugation of maleimide in our related article.
Researchers in industry tend to attach the cytotoxic (anticancer) medicine through these disulfide or non-cleavable thioether linker chemistry to monoclonal antibodies because of the added homogeneity compared to lysine based chemistry (source).
FLIA And Fluorescent Dye-Labeled Antibodies
Fluorescent dyes have the ability to absorb electromagnetic radiation at one wavelength and then emit it at a longer wavelength. For the detection of small amounts of the target compound/metabolite/protein, this Stoke’s shift is critical.
Fluorescently labeled antibodies provide binding specificity and can be utilized for studying localization in FLIA (fluorescence immunoassays). A succinimidyl-ester functional group coupled to a fluorophore core targets primary amines on the antibody to produce a stable covalent connection in fluorescence labeling. The amount of labeling an antibody receives is determined by the number of free primary amine groups and whether the fluorophore has a steric effect on binding specificity, avidity, or affinity.
Some challenges with creating fluorochrome-labeled antibodies are that there might be too few or too many fluorophores, non-specific staining, and loss of antibody-antigen specificity or affinity. Fluorophore-labeled antibodies are employed more for immunohistochemistry and FACS than for quantitative immunoassays because of this (source).
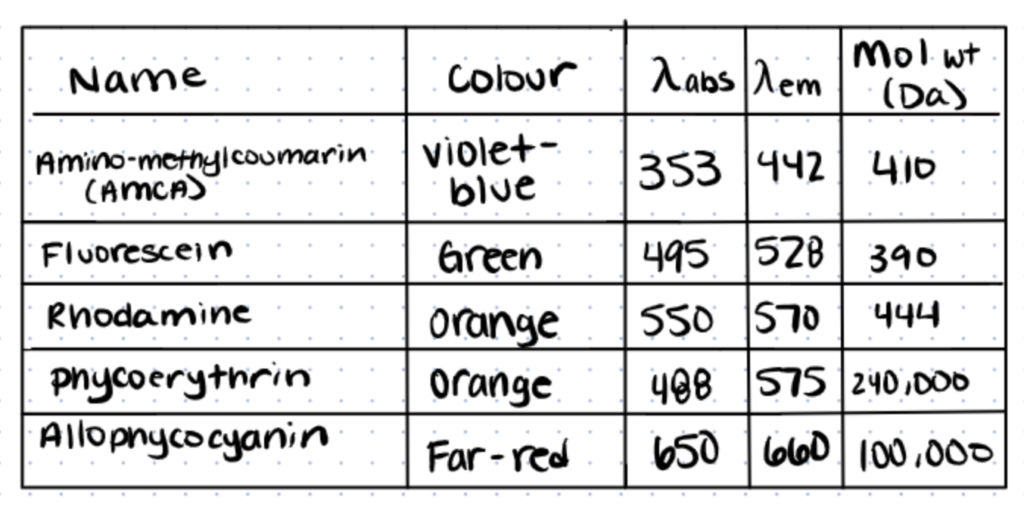
These dyes are the source of most other commercial dyes, including Alexa dyes and Texas Red.
Thiol-modified fluorescent dyes are available in commercial kits. Using an ‘activation’ reagent such as SMCC (succinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate), these dyes can be linked to lysine groups (which contain amines) in a single step. For one step fluorophore labeling, a solution of SMCC in dimethylformamide is incubated with an amine modified fluorophore at a high pH (typically pH 9.0) and an antibody.
The unused fluorescent probe is then removed using a quenching reagent. To eliminate the unconjugated probe, HPLC purification of the labeled antibody is recommended. Rigau M et al, for example, used amine coupling from Thermo Fisher to conjugate an anti-BTN2A1 mAb to Alexa Fluor-647 and a sulfo-SMCC heterobifunctional crosslinker to conjugate an anti-BTN3A antibody to R-phycoerythrin.
HPLC is commonly used to purify, select, and quantify bioconjugation products, read more about this process in our article, bioconjugation characterization.
Antibody Conjugation to Gold Nanoparticles
When compared to native molecules, nanoparticles of many compounds have enhanced activity profiles. Nanoparticle-antibody conjugates can be employed for both therapeutic or diagnostic applications.
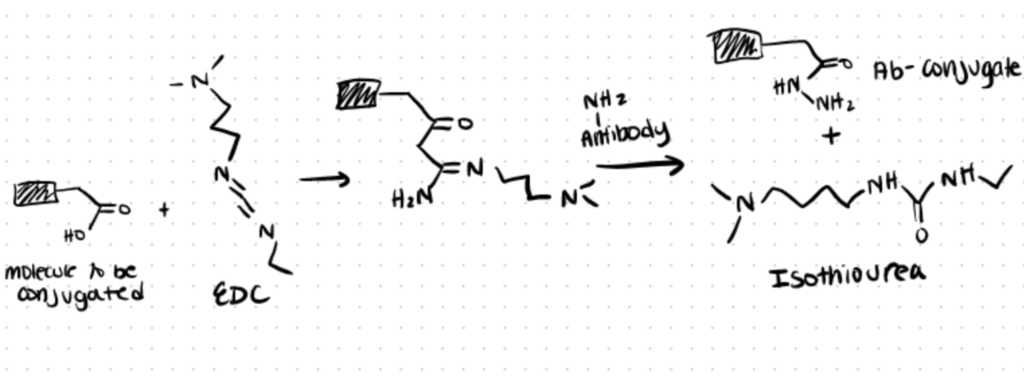
To attach antibodies with gold nanoparticles or colloids, simple EDC/NHS chemistry can be used. EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride) is a common zero-length crosslinker that will crosslink carboxyl groups on nanoparticles with amines of an antibody. It is water-soluble and can easily be neutralized with dilute acid. However, one downside of this approach is that it may lead to non-selective creation of amide bonds around the antibody’s antigen-binding sites and reduce affinity and avidity for the antigen. (source)
There are several other protocols for bioconjugation of nanoparticles that you can find in our linker article.
You can also use EDC/NHS chemistry to attach antibodies to quantum dots to create highly sensitive imaging probes. Here are some step-by-step methods for bioconjugation of quantum dots. In addition, we talk about the creation of an antibody quantum-dot conjugate system through hydrazine bioconjugation.
Other Antibody Conjugates
Antibodies have also been coupled to haptens like biotin, streptavidin, and biomagnetic beads for use in a variety of applications. (source)
You’re probably tired of reading about applications by now, so let’s get into antibody conjugation protocols.
Antibody Conjugation Protocols
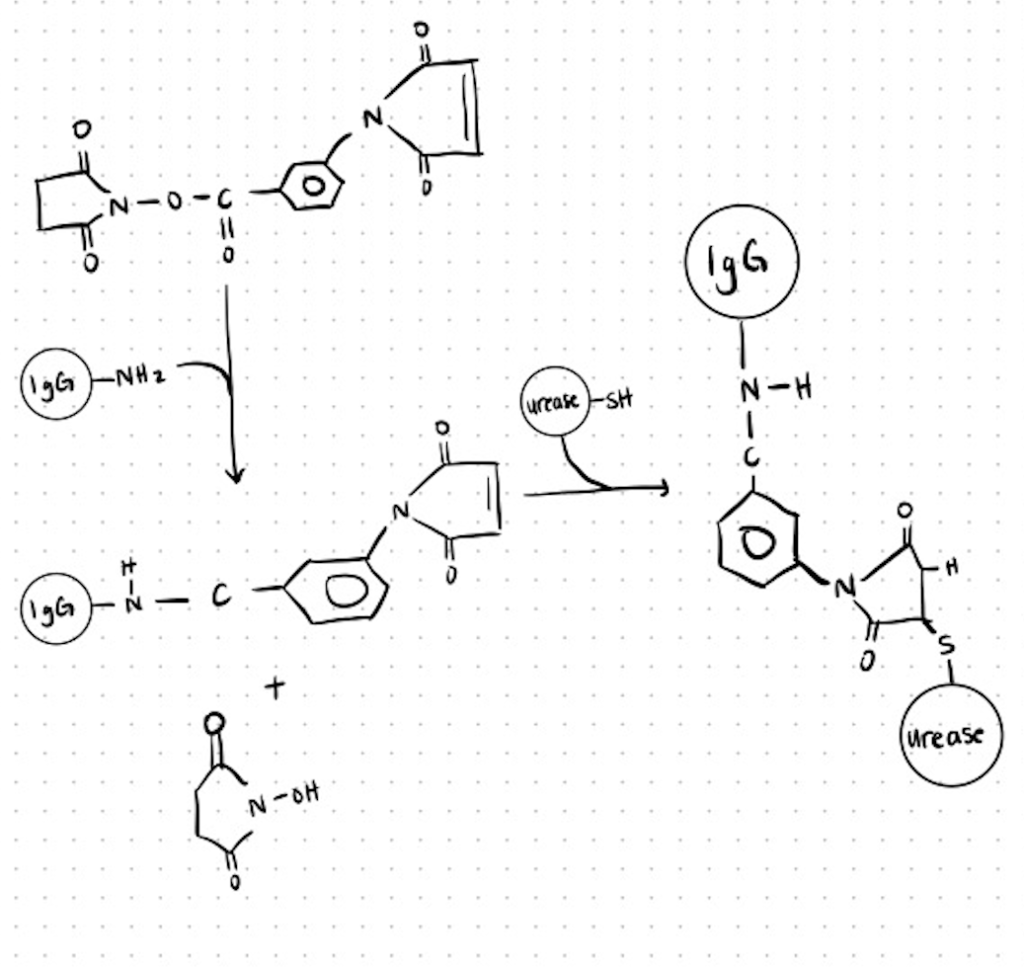
Method 1. Conjugation Of Urease to Antibody (Igg) With m-Maleimidobenzoyl N-Hydroxysuccinimide Ester (Mbs).
This conjugate was used for the detection of human IgM and IgG antibodies against Mycoplasma pneumoniae. You can find more details about the methods utilized and the full protocol here.
Antibodies can be easily attached to other biomolecules like proteins, polymers, and carbohydrates using amines, carboxyls, and even thiols. Use these antibody conjugation kits to attach antibodies with other biomolecules.
Step 1. Desalt Antibody and React with MBS
Overnight at 4°C, dialyze 0.25 ml of 20 mg/ml urease in 0.1 M phosphate buffer against 2 liters 0.1 M phosphate buffer while stirring slowly.
Replace the phosphate buffer in the morning and dialyze for another 2 hours.
Separate the antibody and urease solutions in separate glass tubes from the dialysis membranes. Read the A280 of the antibody solution and dilute to 0.5 mg/ml with phosphate buffer.
In a glass tube, mix 1.5 ml of 0.5 mg/ml dialyzed antibody solution with 0.075 ml MBS/DMF solution (MBS/antibody molar ratio, 1201). Place a magnetic stir bar in the tube and gently stir for 30 minutes at room temperature
Step 2. Fractionate the Conjugate
Pre-equilibrate a PD-10 column with 100 mL phosphate buffer. Collect 0.6-ml fractions after running the column with phosphate buffer. Read fractions A280 and take the first peak that elutes.
Add 0.15 ml of 20 mg/ml urease in 0.1 M phosphatase buffer to the first peak (usually 2.5 to 3.0 ml) (urease/antibody weight ratio, 41).
Stir for 1.5 hours at room temperature under N2 until the solution becomes hazy.
Step 3. Quench Unreacted Conjugates
Dilute 0.143 M 2-mercaptoethanol to 2 mM (0.014 ml mercaptoethanol solution/ml urease-antibody conjugate) and stir for 30 minutes at room temperature.
Dialyze against 2 liters PBS overnight at 4°C. Replace PBS in the morning and dialyze for 4 hours.
Mix with an equal amount of glycerol, split into tiny aliquots and keep at 20°C for up to a year.
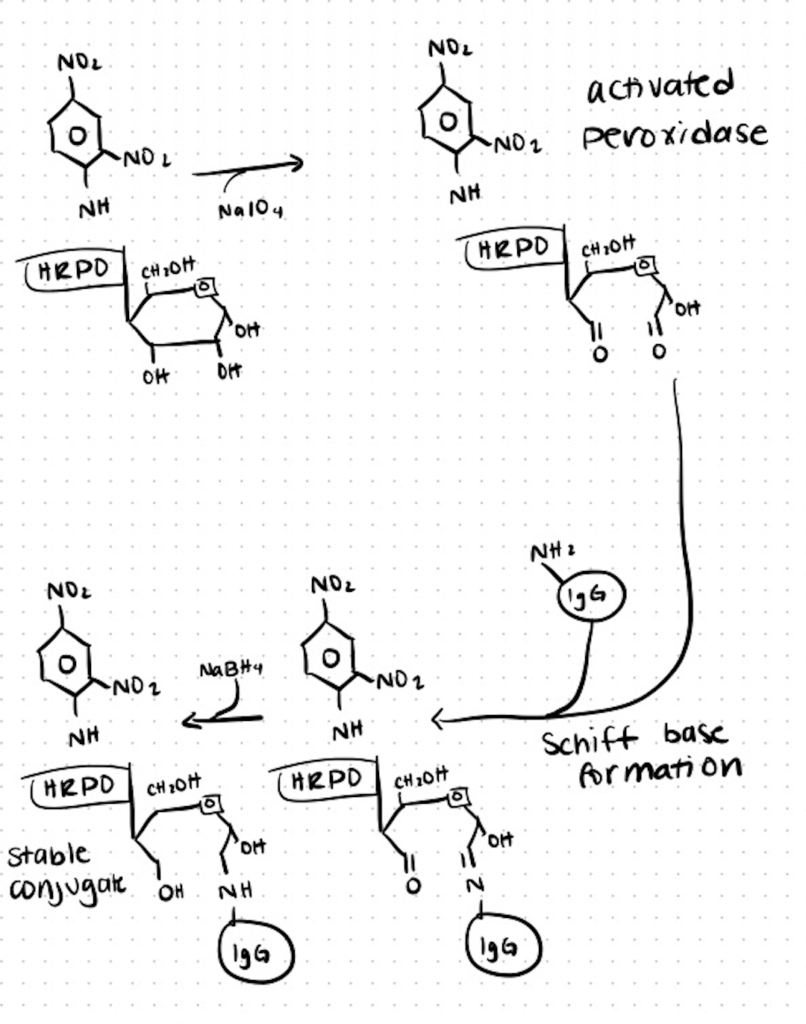
Method 2. Conjugation of Antibodies to Horseradish Peroxidase
One of the most extensively utilized bioreagents in the biological sciences is antibodies coupled with horseradish peroxidase (HRP). Here is a basic method for attaching HRP to a thiolated antibody that may be modified to work with various cross-linkers. Because there are only six of them and their alteration has little effect on enzyme activity, most conjugation methods rely on linking through the lysines on HRP. You can find the full method here at CSH.
Step 1. Add N-Succinimidyl S-acetylthioacetate (SATA) to Antibody
Immediately before usage, dissolve SATA in DMSO to a concentration of 10 mg/mL.
For every milligram of antibody, add 1 milliliter of SATA in a buffer (pH 6.5–7.5).
Combine the ingredients and incubate for 30 minutes at room temperature. The activated antibody should be kept at 4°C until it is needed.
For every 10 volumes of antibody solution, add 1 volume of hydroxylamine buffer to deprotect the thiol groups. Mix the materials together and let the reaction sit at room temperature for 2 hours.
Using a G-25 sizing column, desalt the solution. To avoid disulfide formation, move on to the following step right away.
For each milligram of thiolated antibody, weigh 1.1 mg of HRP. Per milligram of HRP, add 1.1 mg of SMCC. PBS should be diluted to a final concentration of 8 mg/mL HRP. On a rotator, mix for 2 hours.
On a G-25 column, desalt the HRP.
Step 2. Combine the SATA-conjugated HRP with the Activated Antibody
Combine the HRP with the thiolated antibody right away, using 1 mg of HRP per milligram of antibody. Incubate at 4°C for 16–20 hours with mixing.
Dialyze for 2 hours at room temperature against three changes of PBS. Ion exchange or affinity chromatography may be required to separate the free enzyme from the conjugate.
Using filtration, sterilize the enzyme–antibody conjugate solution. At 4°C, store for up to 30 days. Lyophilize and store at 70°C for up to a year for longer storage. Before utilizing in the appropriate application, immediately reconstitute.
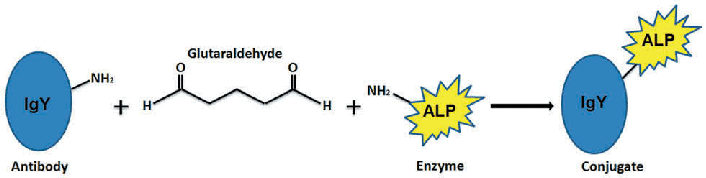
Method 3. Conjugation of Alkaline Phosphatase to antibodies
The homobifunctional reagent glutaraldehyde, which interacts with amino groups in two proteins, is used to link alkaline phosphatase to IgG antibody in a one-step process. The operation is straightforward to carry out and involves only a few pieces of equipment. You can find the full protocol in Current Techniques in Molecular Biology.
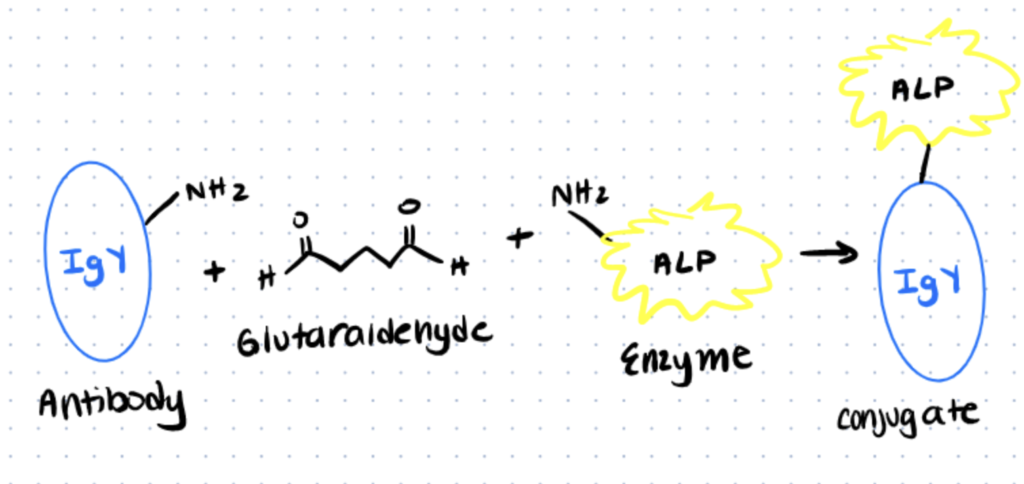
Step 1. Dialysis of Antibody Solution
Dialyze 1 mg/ml antibody solution overnight at 4°C in 2 liters of 0.1 M phosphate buffer, pH 6.8, stirring gently.
Place the antibody solution in a tube after removing it from the dialysis membrane. Read A280 and dilute to 3 mg/ml in PBS
In a 1.5-ml microcentrifuge tube, mix 100 ul dialyzed antibody solution with 90 ul alkaline phosphatase (10 mg/ml stock).
Step 2. Reaction with Glutaraldehyde
Gently stir in 5 mL of 25 percent glutaraldehyde. Allow cooling to room temperature.
Step 3. Sample Collection
At time 0, 5, 10, 15, 30, 60, and 120 minutes, take 25 μl samples and put them in separate 1.5-ml microcentrifuge tubes. To each sample, add 125 μl PBS, then 1.1 ml Tris/ovalbumin solution. Keep each sample refrigerated until the time course is finished.
Step 4. Storage of Conjugates
Add sodium azide to 0.1 percent and keep the conjugate at 4°C for up to a year, shielded from light. Alternatively, add an equivalent volume of glycerol to the conjugates and keep them at 20°C for 1 year.
